9. Chapter 8 (Pine): Curve Fitting Solutions¶
\({\large\bf 1.}\) We linearize the equation \(V(t)=V_0e^{\Gamma t}\) by taking the logarithm: \(\ln V = \ln V_0 + \Gamma t\). Comparing with the equation for a straight line \(Y = A + BX\), we see that
\(\bf{(a)}\) & \(\bf{(c)}\) There are two parts to this problem: (1) writing the fitting function with \(\chi^2\) weighting and (2) transforming the data to linear form so that it can be fit to an exponential.
The first part is done with the function LineFitWt(x, y, sig). There is also an ancillary function rechisq(x, y, dy, slope, yint) that calcuates the reduced chi-squared \(\chi_r^2\) for a particular set of data & fitting parameters.
The second part involves transforming the data and its uncertainties. This is done following the procedure described in Introduction to Python for Science (by Pine) in \(\S 8.1.1\).
[1]:
import numpy as np
import matplotlib.pyplot as plt
def LineFitWt(x, y, sig):
"""
Returns slope and y-intercept of weighted linear fit to
(x,y) data set.
Inputs: x and y data array and uncertainty array (unc)
for y data set.
Outputs: slope and y-intercept of best fit to data and
uncertainties of slope & y-intercept.
"""
sig2 = sig ** 2
norm = (1.0 / sig2).sum()
xhat = (x / sig2).sum() / norm
yhat = (y / sig2).sum() / norm
slope = ((x - xhat) * y / sig2).sum() / ((x - xhat) * x / sig2).sum()
yint = yhat - slope * xhat
sig2_slope = 1.0 / ((x - xhat) * x / sig2).sum()
sig2_yint = sig2_slope * (x * x / sig2).sum() / norm
return slope, yint, np.sqrt(sig2_slope), np.sqrt(sig2_yint)
def redchisq(x, y, dy, slope, yint):
chisq = (((y - yint - slope * x) / dy) ** 2).sum()
return chisq / float(x.size - 2)
# Read data from data file
t, V, dV = np.loadtxt("data/RLcircuit.txt", skiprows=2, unpack=True)
########## Code to tranform & fit data starts here ##########
# Transform data and parameters from ln V = ln V0 - Gamma t
# to linear form: Y = A + B*X, where Y = ln V, X = t, dY = dV/V
X = t # transform t data for fitting (not needed as X=t)
Y = np.log(V) # transform N data for fitting
dY = dV / V # transform uncertainties for fitting
# Fit transformed data X, Y, dY to obtain fitting parameters
# B & A. Also returns uncertainties dA & dB in B & A
B, A, dB, dA = LineFitWt(X, Y, dY)
# Return reduced chi-squared
redchisqr = redchisq(X, Y, dY, B, A)
# Determine fitting parameters for original exponential function
# N = N0 exp(-Gamma t) ...
V0 = np.exp(A)
Gamma = -B
# ... and their uncertainties
dV0 = V0 * dA
dGamma = dB
###### Code to plot transformed data and fit starts here ######
# Create line corresponding to fit using fitting parameters
# Only two points are needed to specify a straight line
Xext = 0.05 * (X.max() - X.min())
Xfit = np.array([X.min() - Xext, X.max() + Xext]) # smallest & largest X points
Yfit = B * Xfit + A # generates Y from X data &
# fitting function
plt.errorbar(X, Y, dY, fmt="b^")
plt.plot(Xfit, Yfit, "c-", zorder=-1)
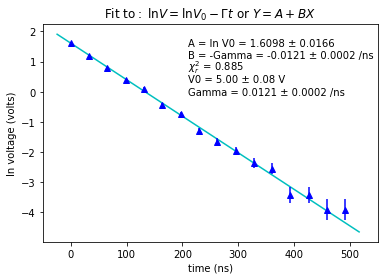
plt.title(r"$\mathrm{Fit\ to:}\ \ln V = \ln V_0-\Gamma t$ or $Y = A + BX$")
plt.xlabel("time (ns)")
plt.ylabel("ln voltage (volts)")
plt.xlim(-50, 550)
plt.text(210, 1.5, u"A = ln V0 = {0:0.4f} \xb1 {1:0.4f}".format(A, dA))
plt.text(210, 1.1, u"B = -Gamma = {0:0.4f} \xb1 {1:0.4f} /ns".format(B, dB))
plt.text(210, 0.7, "$\chi_r^2$ = {0:0.3f}".format(redchisqr))
plt.text(210, 0.3, u"V0 = {0:0.2f} \xb1 {1:0.2f} V".format(V0, dV0))
plt.text(210, -0.1, u"Gamma = {0:0.4f} \xb1 {1:0.4f} /ns".format(Gamma, dGamma))
plt.show()
plt.savefig("RLcircuit.pdf")
<Figure size 432x288 with 0 Axes>
\(\bf{(b)}\) The value of \(\chi_r^2\) returned by the fitting routine is \(0.885\), which is near 1, so it seem that the error bars are about right and an exponential is a good model for the data.
\({\bf (d)}\) Starting from \(\Gamma = R/L\) and assuming negligible uncertainty in \(R\), we have
Here are the calculations:
[2]:
R = 10.0e3
Gamma *= 1.0e9 # convert Gamma from 1/ns to 1/s
L = R / Gamma
print("L = {0:0.2e} henry".format(L))
dGamma *= 1.0e9 # convert dGamma from 1/ns to 1/s
dL = L * (dGamma / Gamma)
print("dL = {0:0.1e} henry".format(dL))
L = 8.24e-04 henry
dL = 1.1e-05 henry
\({\large\bf 2.}\) Here we want to use a linear fitting routine (\(Y = A + BX\)) to fit a power law model
where \(K\) and \(p\) are fitting parameters. We transform the equation by taking the logarithm of both sides, which gives
Thus, identifying the transformed variables as
and the \(y\)-intercept and slope and are given by \(A=\ln K\) and \(B=p\), respectively.
The uncertainties in \(y\) are related to those in \(m\) by
The uncertainties in the fitting paramters follow from \(K=e^A\) and \(p=B\):
These transformations are implemented in the code below. We use the same fitting routine used in Problem 1 above.
[3]:
import numpy as np
import matplotlib.pyplot as plt
def LineFitWt(x, y, sig):
"""
Returns slope and y-intercept of weighted linear fit to
(x,y) data set.
Inputs: x and y data array and uncertainty array (unc)
for y data set.
Outputs: slope and y-intercept of best fit to data and
uncertainties of slope & y-intercept.
"""
sig2 = sig ** 2
norm = (1.0 / sig2).sum()
xhat = (x / sig2).sum() / norm
yhat = (y / sig2).sum() / norm
slope = ((x - xhat) * y / sig2).sum() / ((x - xhat) * x / sig2).sum()
yint = yhat - slope * xhat
sig2_slope = 1.0 / ((x - xhat) * x / sig2).sum()
sig2_yint = sig2_slope * (x * x / sig2).sum() / norm
return slope, yint, np.sqrt(sig2_slope), np.sqrt(sig2_yint)
def redchisq(x, y, dy, slope, yint):
chisq = (((y - yint - slope * x) / dy) ** 2).sum()
return chisq / float(x.size - 2)
# Read data from data file
n, m, dm = np.loadtxt("data/Mass.txt", skiprows=4, unpack=True)
########## Code to tranform & fit data starts here ##########
# Transform data and parameters to linear form: Y = A + B*X
X = np.log(m) # transform t data for fitting
Y = np.log(n) # transform N data for fitting
dY = dm / m # transform uncertainties for fitting
# Fit transformed data X, Y, dY to obtain fitting parameters
# B & A. Also returns uncertainties dA & dB in B & A
B, A, dB, dA = LineFitWt(X, Y, dY)
# Return reduced chi-squared
redchisqr = redchisq(X, Y, dY, B, A)
# Determine fitting parameters for original exponential function
# N = N0 exp(-Gamma t) ...
p = B
K = np.exp(A)
# ... and their uncertainties
dp = dB
dK = np.exp(A) * dA
###### Code to plot transformed data and fit starts here ######
# Create line corresponding to fit using fitting parameters
# Only two points are needed to specify a straight line
Xext = 0.05 * (X.max() - X.min())
Xfit = np.array([X.min() - Xext, X.max() + Xext])
Yfit = B * Xfit + A # generates Y from X data &
# fitting function
plt.errorbar(X, Y, dY, fmt="gs")
plt.plot(Xfit, Yfit, "k-", zorder=-1)
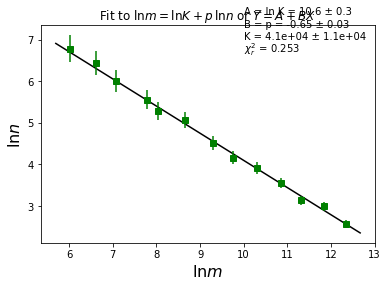
plt.title(r"Fit to $\ln m=\ln K + p\, \ln n$ or $Y=A+BX$")
plt.xlabel(r"$\ln m$", fontsize=16)
plt.ylabel(r"$\ln n$", fontsize=16)
plt.text(10, 7.6, u"A = ln K = {0:0.1f} \xb1 {1:0.1f}".format(A, dA))
plt.text(10, 7.3, u"B = p = {0:0.2f} \xb1 {1:0.2f}".format(B, dB))
plt.text(10, 7.0, u"K = {0:0.1e} \xb1 {1:0.1e}".format(K, dK))
plt.text(10, 6.7, "$\chi_r^2$ = {0:0.3f}".format(redchisqr))
plt.show()
plt.savefig("Mass.pdf")
<Figure size 432x288 with 0 Axes>
\({\large\bf 3.}\) (a)
[4]:
import numpy as np
import matplotlib.pyplot as plt
# define function to calculate reduced chi-squared
def RedChiSqr(func, x, y, dy, params):
resids = y - func(x, *params)
chisq = ((resids / dy) ** 2).sum()
return chisq / float(x.size - params.size)
# define fitting function
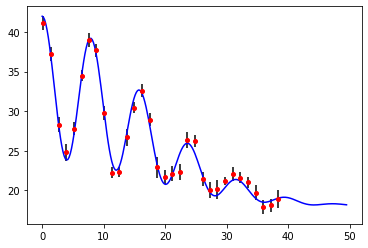
def oscDecay(t, A, B, C, tau, omega):
y = A * (1.0 + B * np.cos(omega * t)) * np.exp(-0.5 * t * t / (tau * tau)) + C
return y
# read in spectrum from data file
t, decay, unc = np.loadtxt("data/oscDecayData.txt", skiprows=4, unpack=True)
# initial values for fitting parameters (guesses)
A0 = 15.0
B0 = 0.6
C0 = 1.2 * A0
tau0 = 16.0
omega0 = 2.0 * np.pi / 8.0 # period of oscillations in data is about 8
# plot data and fit with estimated fitting parameters
tFit = np.linspace(0.0, 49.5, 250)
plt.plot(tFit, oscDecay(tFit, A0, B0, C0, tau0, omega0), "b-")
plt.errorbar(t, decay, yerr=unc, fmt="or", ecolor="black", ms=4)
plt.show()
[5]:
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec # for unequal plot boxes
import scipy.optimize
# define function to calculate reduced chi-squared
def RedChiSqr(func, x, y, dy, params):
resids = y - func(x, *params)
chisq = ((resids / dy) ** 2).sum()
return chisq / float(x.size - params.size)
# define fitting function
def oscDecay(t, A, B, C, tau, omega):
y = A * (1.0 + B * np.cos(omega * t)) * np.exp(-0.5 * t * t / (tau * tau)) + C
return y
# read in spectrum from data file
t, decay, unc = np.loadtxt("data/oscDecayData.txt", skiprows=4, unpack=True)
# initial values for fitting parameters (guesses)
A0 = 15.0
B0 = 0.6
C0 = 1.2 * A0
tau0 = 16.0
omega0 = 2.0 * np.pi / 8.0
# fit data using SciPy's Levenberg-Marquart method
nlfit, nlpcov = scipy.optimize.curve_fit(
oscDecay, t, decay, p0=[A0, B0, C0, tau0, omega0], sigma=unc
)
# calculate reduced chi-squared
rchi = RedChiSqr(oscDecay, t, decay, unc, nlfit)
# create fitting function from fitted parameters
A, B, C, tau, omega = nlfit
t_fit = np.linspace(0.0, 50.0, 512)
d_fit = oscDecay(t_fit, A, B, C, tau, omega)
# Create figure window to plot data
fig = plt.figure(1, figsize=(8, 8)) # extra length for residuals
gs = gridspec.GridSpec(2, 1, height_ratios=[6, 2])
# Top plot: data and fit
ax1 = fig.add_subplot(gs[0])
ax1.plot(t_fit, d_fit)
ax1.errorbar(t, decay, yerr=unc, fmt="or", ecolor="black", ms=4)
ax1.set_xlabel("time (ms)")
ax1.set_ylabel("decay (arb units)")
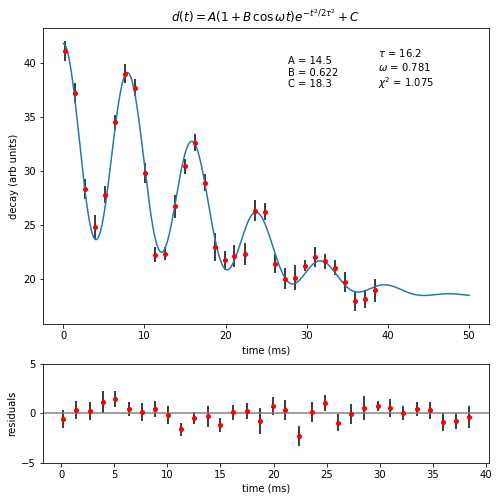
ax1.text(
0.55,
0.8,
"A = {0:0.1f}\nB = {1:0.3f}\nC = {2:0.1f}".format(A, B, C),
transform=ax1.transAxes,
)
ax1.text(
0.75,
0.8,
"$\\tau$ = {0:0.1f}\n$\omega$ = {1:0.3f}\n$\chi^2$ = {2:0.3f}".format(
tau, omega, rchi
),
transform=ax1.transAxes,
)
ax1.set_title("$d(t) = A (1+B\,\cos\,\omega t) e^{-t^2/2\\tau^2} + C$")
# Bottom plot: residuals
resids = decay - oscDecay(t, A, B, C, tau, omega)
ax2 = fig.add_subplot(gs[1])
ax2.axhline(color="gray")
ax2.errorbar(t, resids, yerr=unc, ecolor="black", fmt="ro", ms=4)
ax2.set_xlabel("time (ms)")
ax2.set_ylabel("residuals")
ax2.set_ylim(-5, 5)
yticks = (-5, 0, 5)
ax2.set_yticks(yticks)
plt.savefig("FitOscDecay.pdf")
plt.show()
[6]:
# initial values for fitting parameters (guesses)
A0 = 15.0
B0 = 0.6
C0 = 1.2 * A0
tau0 = 16.0
omega0 = 3.0 * 0.781
# fit data using SciPy's Levenberg-Marquart method
nlfit, nlpcov = scipy.optimize.curve_fit(
oscDecay, t, decay, p0=[A0, B0, C0, tau0, omega0], sigma=unc
)
# calculate reduced chi-squared
rchi = RedChiSqr(oscDecay, t, decay, unc, nlfit)
# create fitting function from fitted parameters
A, B, C, tau, omega = nlfit
t_fit = np.linspace(0.0, 50.0, 512)
d_fit = oscDecay(t_fit, A, B, C, tau, omega)
# Create figure window to plot data
fig = plt.figure(1, figsize=(8, 6))
# Top plot: data and fit
ax1 = fig.add_subplot(111)
ax1.plot(t_fit, d_fit)
ax1.errorbar(t, decay, yerr=unc, fmt="or", ecolor="black", ms=4)
ax1.set_xlabel("time (ms)")
ax1.set_ylabel("decay (arb units)")
ax1.text(
0.55,
0.8,
"A = {0:0.1f}\nB = {1:0.3f}\nC = {2:0.1f}".format(A, B, C),
transform=ax1.transAxes,
)
ax1.text(
0.75,
0.8,
"$\\tau$ = {0:0.1f}\n$\omega$ = {1:0.3f}\n$\chi^2$ = {2:0.3f}".format(
tau, omega, rchi
),
transform=ax1.transAxes,
)
ax1.set_title("$d(t) = A (1+B\,\cos\,\omega t) e^{-t^2/2\\tau^2} + C$")
plt.show()
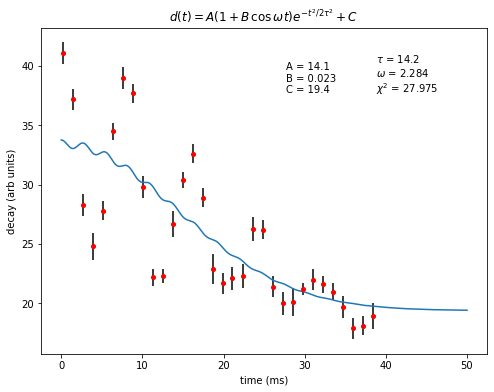
The program finds the optimal values for all the fitting paramters again
[7]:
# initial values for fitting parameters (guesses)
A0 = 15.0
B0 = 0.6
C0 = 1.2 * A0
tau0 = 16.0
omega0 = 2.0 * np.pi / 8.0
# fit data using SciPy's Levenberg-Marquardt method
nlfit, nlpcov = scipy.optimize.curve_fit(
oscDecay, t, decay, p0=[A0, B0, C0, tau0, omega0], sigma=unc
)
# unpack optimal values of fitting parameters from nlfit
A, B, C, tau, omega = nlfit
# calculate reduced chi square for different values around the optimal
omegaArray = np.linspace(0.05, 2.95, 256)
redchiArray = np.zeros(omegaArray.size)
for i in range(omegaArray.size):
nlfit = np.array([A, B, C, tau, omegaArray[i]])
redchiArray[i] = RedChiSqr(oscDecay, t, decay, unc, nlfit)
plt.figure(figsize=(8, 4))
plt.plot(omegaArray, redchiArray)
plt.xlabel("$\omega$")
plt.ylabel("$\chi_r^2$")
plt.savefig("VaryChiSq.pdf")
plt.show()
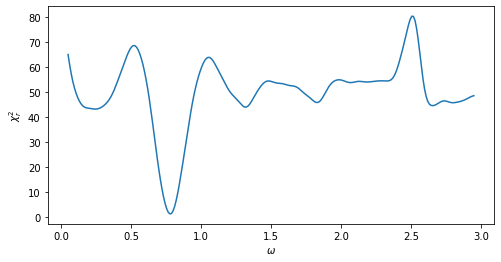
[ ]: