Table of Contents
1 02: xarray, netcdf and zarr
1.1 The current defacto standard in atmos/ocean science
1.2 Some challenges with netcdf
1.3 create an xarray
1.4 Download toy model data
1.5 Sort in numeric order
1.6 Make an xarray
1.7 Dump to a zarr file
02: xarray, netcdf and zarr¶
Motivation: how you store your data can an enormous effect on performance.
Four issues:
- Compression vs. cpu time to uncompress/compress
- Multithreaded read/writes
- Performance for cloud storage (Amazon, Google Compute, Azure)
- Throttling data reads to fit in available memory and avoid swapping (“chunking”)
The current defacto standard in atmos/ocean science¶
Some challenges with netcdf¶
create an xarray¶
[1]:
import glob
import numpy as np
import pdb
[2]:
#!conda install -y xarray
[3]:
import context
from westgrid.data_read import download
******************************
context imported. Front of path:
/Users/phil/repos/parallel_python_course/notebooks
******************************
Download toy model data¶
[4]:
#get 10 files, each is the same timestep for a member of a
#10 member ensemble
import numpy as np
root='http://clouds.eos.ubc.ca/~phil/docs/atsc500/small_les'
for i in range(10):
the_name=f'mar12014_{(i+1):d}_15600.nc'
print(the_name)
url='{}/{}'.format(root,the_name)
download(the_name,root=root)
mar12014_1_15600.nc
trying http://clouds.eos.ubc.ca/~phil/docs/atsc500/small_les/mar12014_1_15600.nc
writing to: mar12014_1_15600.nc
mar12014_1_15600.nc already exists
and is 1455454 bytes
will not overwrite
mar12014_2_15600.nc
trying http://clouds.eos.ubc.ca/~phil/docs/atsc500/small_les/mar12014_2_15600.nc
writing to: mar12014_2_15600.nc
mar12014_2_15600.nc already exists
and is 1455454 bytes
will not overwrite
mar12014_3_15600.nc
trying http://clouds.eos.ubc.ca/~phil/docs/atsc500/small_les/mar12014_3_15600.nc
writing to: mar12014_3_15600.nc
mar12014_3_15600.nc already exists
and is 1455454 bytes
will not overwrite
mar12014_4_15600.nc
trying http://clouds.eos.ubc.ca/~phil/docs/atsc500/small_les/mar12014_4_15600.nc
writing to: mar12014_4_15600.nc
mar12014_4_15600.nc already exists
and is 1455454 bytes
will not overwrite
mar12014_5_15600.nc
trying http://clouds.eos.ubc.ca/~phil/docs/atsc500/small_les/mar12014_5_15600.nc
writing to: mar12014_5_15600.nc
mar12014_5_15600.nc already exists
and is 1455454 bytes
will not overwrite
mar12014_6_15600.nc
trying http://clouds.eos.ubc.ca/~phil/docs/atsc500/small_les/mar12014_6_15600.nc
writing to: mar12014_6_15600.nc
mar12014_6_15600.nc already exists
and is 1455454 bytes
will not overwrite
mar12014_7_15600.nc
trying http://clouds.eos.ubc.ca/~phil/docs/atsc500/small_les/mar12014_7_15600.nc
writing to: mar12014_7_15600.nc
mar12014_7_15600.nc already exists
and is 1455454 bytes
will not overwrite
mar12014_8_15600.nc
trying http://clouds.eos.ubc.ca/~phil/docs/atsc500/small_les/mar12014_8_15600.nc
writing to: mar12014_8_15600.nc
mar12014_8_15600.nc already exists
and is 1455454 bytes
will not overwrite
mar12014_9_15600.nc
trying http://clouds.eos.ubc.ca/~phil/docs/atsc500/small_les/mar12014_9_15600.nc
writing to: mar12014_9_15600.nc
mar12014_9_15600.nc already exists
and is 1455454 bytes
will not overwrite
mar12014_10_15600.nc
trying http://clouds.eos.ubc.ca/~phil/docs/atsc500/small_les/mar12014_10_15600.nc
writing to: mar12014_10_15600.nc
mar12014_10_15600.nc already exists
and is 1455454 bytes
will not overwrite
[5]:
#get 10 files, each is the same timestep for a member of a
#10 member ensemble
import numpy as np
root=Path().resolve()
the_files=root.glob('mar12*nc')
the_files=list(the_files)
the_files
[5]:
[PosixPath('/Users/phil/repos/parallel_python_course/notebooks/mar12014_2_15600.nc'),
PosixPath('/Users/phil/repos/parallel_python_course/notebooks/mar12014_3_15600.nc'),
PosixPath('/Users/phil/repos/parallel_python_course/notebooks/mar12014_10_15600.nc'),
PosixPath('/Users/phil/repos/parallel_python_course/notebooks/mar12014_8_15600.nc'),
PosixPath('/Users/phil/repos/parallel_python_course/notebooks/mar12014_9_15600.nc'),
PosixPath('/Users/phil/repos/parallel_python_course/notebooks/mar12014_5_15600.nc'),
PosixPath('/Users/phil/repos/parallel_python_course/notebooks/mar12014_4_15600.nc'),
PosixPath('/Users/phil/repos/parallel_python_course/notebooks/mar12014_1_15600.nc'),
PosixPath('/Users/phil/repos/parallel_python_course/notebooks/mar12014_6_15600.nc'),
PosixPath('/Users/phil/repos/parallel_python_course/notebooks/mar12014_7_15600.nc')]
Sort in numeric order¶
[6]:
import glob
import xarray
from matplotlib import pyplot as plt
import numpy as np
def sort_name(pathname):
"""
sort the filenames so '10' sorts
last by converting to integers
"""
str_name=str(pathname.name)
front, number, back = str_name.split('_')
return int(number)
Make an xarray¶
Now use xarray to stitch together the 10 ensemble members along a new “virtual dimenstion”. The variable “ds” is an xray dataset, which controls the reads/writes from the 10 netcdf files
[7]:
the_files.sort(key=sort_name)
#
# put the 10 ensembles together along a new "ens" dimension
# using an xray multi-file dataset
#
ds = xarray.open_mfdataset(the_files, engine='netcdf4', concat_dim='ens')
# dump the structure
print(ds)
#
# 3-d ensemble average for temp
#
x = ds['x']
y = ds['y']
z = ds['z']
temp = ds['TABS']
mean_temp = temp[:, :, :, :].mean(dim='ens')
#
# same for velocity
#
wvel = ds['W']
mean_w = wvel[:, :, :, :].mean(dim='ens')
#
# now look at the perturbation fields for one ensemble member
#
wvelprime = wvel[0, :, :, :] - mean_w
Tprime = temp[0, :, :, :] - mean_temp
flux_prime = wvelprime * Tprime
flux_profile = flux_prime.mean(dim='x').mean(dim='y')
keep_dict = dict(flux_prof=flux_profile, flux_prime=flux_prime.values,
wvelprime=wvelprime.values, Tprime=Tprime.values, x=x, y=y, z=z)
<xarray.Dataset>
Dimensions: (ens: 10, time: 1, x: 50, y: 40, z: 30)
Coordinates:
* x (x) float32 0.0 25.0 50.0 75.0 100.0 125.0 150.0 175.0 200.0 ...
* y (y) float32 0.0 25.0 50.0 75.0 100.0 125.0 150.0 175.0 200.0 ...
* z (z) float32 25.0 50.0 75.0 100.0 125.0 150.0 175.0 200.0 205.0 ...
* time (time) float32 169.18056
Dimensions without coordinates: ens
Data variables:
p (ens, z) float32 dask.array<shape=(10, 30), chunksize=(1, 30)>
U (ens, z, y, x) float32 dask.array<shape=(10, 30, 40, 50), chunksize=(1, 30, 40, 50)>
V (ens, z, y, x) float32 dask.array<shape=(10, 30, 40, 50), chunksize=(1, 30, 40, 50)>
W (ens, z, y, x) float32 dask.array<shape=(10, 30, 40, 50), chunksize=(1, 30, 40, 50)>
PP (ens, z, y, x) float32 dask.array<shape=(10, 30, 40, 50), chunksize=(1, 30, 40, 50)>
TABS (ens, z, y, x) float32 dask.array<shape=(10, 30, 40, 50), chunksize=(1, 30, 40, 50)>
QV (ens, z, y, x) float32 dask.array<shape=(10, 30, 40, 50), chunksize=(1, 30, 40, 50)>
Dump to a zarr file¶
[8]:
ds.to_zarr('zarr_dir','w')
[8]:
<xarray.backends.zarr.ZarrStore at 0x312a84f28>
[9]:
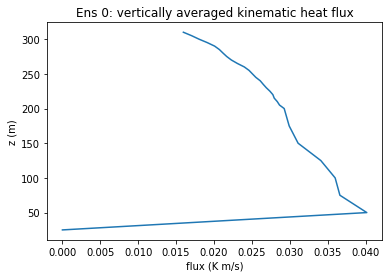
fig1, ax1 = plt.subplots(1, 1)
ax1.plot('flux_prof', 'z', data = keep_dict)
ax1.set(title='Ens 0: vertically averaged kinematic heat flux',
ylabel='z (m)', xlabel='flux (K m/s)')
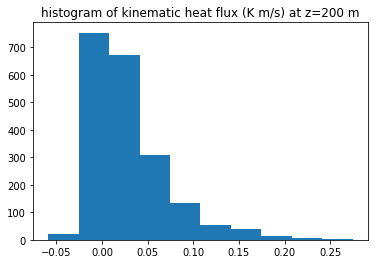
fig2, ax2 = plt.subplots(1, 1)
z200 = np.searchsorted(keep_dict['z'], 200)
flux_200 = keep_dict['flux_prime'][z200,:,:].flat
ax2.hist(flux_200)
ax2.set(title='histogram of kinematic heat flux (K m/s) at z=200 m')
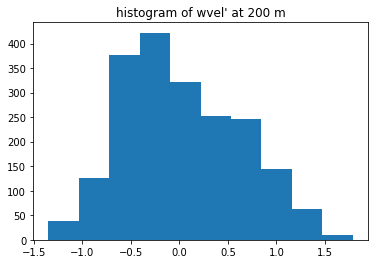
fig3, ax3 = plt.subplots(1, 1)
wvel200=keep_dict['wvelprime'][z200,:,:].flat
ax3.hist(wvel200)
ax3.set(title="histogram of wvel' at 200 m")
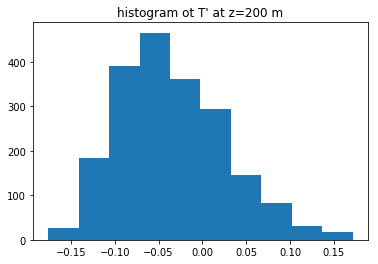
fig4, ax4 = plt.subplots(1, 1)
Tprime200=keep_dict['Tprime'][z200, :, :].flat
ax4.hist(Tprime200)
ax4.set(title="histogram ot T' at z=200 m");