Table of Contents
1 03 - Using dask and zarr for multithreaded input/output
1.1 zarr
1.2 dask
1.2.1 Example, write and read zarr arrays using multiple threads
1.2.2 Create 230 Mbytes of fake data
1.2.3 Copy to a zarr file on disk, using multiple threads
1.2.4 Add some attributes
1.2.5 Create an array of zeros – note that compression shrinks it from 230 Mbytes to 321 bytes
1.2.6 copy input to output using chunks
1.2.7 Create a dask array from a zarr disk file
1.2.8 The following calculation uses numpy, so it releases the GIL
1.2.9 Note that result hasn’t been computed yet
1.2.10 Now do the calculation
1.2.11 You can evaluate your own functions on dask arrays
conda install zarr
conda install dask
conda install pytz #for our timestamp timezone
[1]:
import numpy as np
import zarr
import time
import datetime
import pytz
from zarr.util import human_readable_size
import dask
import dask.array as da
from dask.diagnostics import Profiler, ResourceProfiler, CacheProfiler
from dask.diagnostics.profile_visualize import visualize
03 - Using dask and zarr for multithreaded input/output¶
- Many python programmers use hdf5 through either the h5py or pytables modules to store large dense arrays. One drawback of the hdf5 implementation is that it is basically single-threaded, that is only one core can read or write to a dataset at any one time. Multithreading makes data compression much more attractive, because data sections can be decompressing/compressing simultaneouly while the data is read/written.
zarr¶
- zarr keeps the h5py interface (which is similar to numpy’s), but allows different choices for file compression and is fully multithreaded. See Alistair Miles original blog entry for a discussion of the motivation behind zarr.
dask¶
- dask is a Python library that implements lazy data structures (array, dataframe, bag) and a clever thread/process scheduler. It integrates with zarr to allow calculations on datasets that don’t fit into core memory, either in a single node or across a cluster.
Example, write and read zarr arrays using multiple threads¶
Create 230 Mbytes of fake data¶
[2]:
wvel_data = np.random.normal(2000, 1000, size=[8000,7500]).astype(np.float32)
human_readable_size(wvel_data.nbytes)
[2]:
'228.9M'
Copy to a zarr file on disk, using multiple threads¶
[3]:
item='disk1_data'
store = zarr.DirectoryStore(item)
group=zarr.hierarchy.group(store=store,overwrite=True,synchronizer=zarr.ThreadSynchronizer())
the_var='wvel'
out_zarr1=group.zeros(the_var,shape=wvel_data.shape,dtype=wvel_data.dtype,chunks=[2000,7500])
out_zarr1[...]=wvel_data[...]
Add some attributes¶
[4]:
now=datetime.datetime.now(pytz.UTC)
timestamp= int(now.strftime('%s'))
out_zarr1.attrs['history']='written for practice'
out_zarr1.attrs['creation_date']=str(now)
out_zarr1.attrs['gmt_timestap']=timestamp
out_zarr1
[4]:
<zarr.core.Array '/wvel' (8000, 7500) float32>
Create an array of zeros – note that compression shrinks it from 230 Mbytes to 321 bytes¶
[5]:
a2 = np.zeros([8000,7500],dtype=np.float32)
item='disk2_data'
store = zarr.DirectoryStore(item)
group=zarr.hierarchy.group(store=store,overwrite=True,synchronizer=zarr.ThreadSynchronizer())
the_var='wvel'
out_zarr2=group.zeros(the_var,shape=a2.shape,dtype=a2.dtype,chunks=[2000,7500])
out_zarr2
[5]:
<zarr.core.Array '/wvel' (8000, 7500) float32>
copy input to output using chunks¶
[6]:
item='disk2_data'
store = zarr.DirectoryStore(item)
group=zarr.hierarchy.group(store=store,overwrite=True,synchronizer=zarr.ThreadSynchronizer())
the_var='wvel'
out_zarr=group.empty(the_var,shape=wvel_data.shape,dtype=wvel_data.dtype,chunks=[2000,7500])
out_zarr2[...]=out_zarr1[...]
out_zarr2
[6]:
<zarr.core.Array '/wvel' (8000, 7500) float32>
Create a dask array from a zarr disk file¶
[7]:
da_input = da.from_array(out_zarr2, chunks=out_zarr1.chunks)
da_input
[7]:
dask.array<array, shape=(8000, 7500), dtype=float32, chunksize=(2000, 7500)>
The following calculation uses numpy, so it releases the GIL¶
[8]:
result=(da_input**2. + da_input**3.).mean(axis=0)
result
[8]:
dask.array<mean_agg-aggregate, shape=(7500,), dtype=float32, chunksize=(7500,)>
Note that result hasn’t been computed yet¶
Here is a graph of how the calculation will be split among 4 threads
[9]:
from dask.dot import dot_graph
dot_graph(result.dask)
[9]:
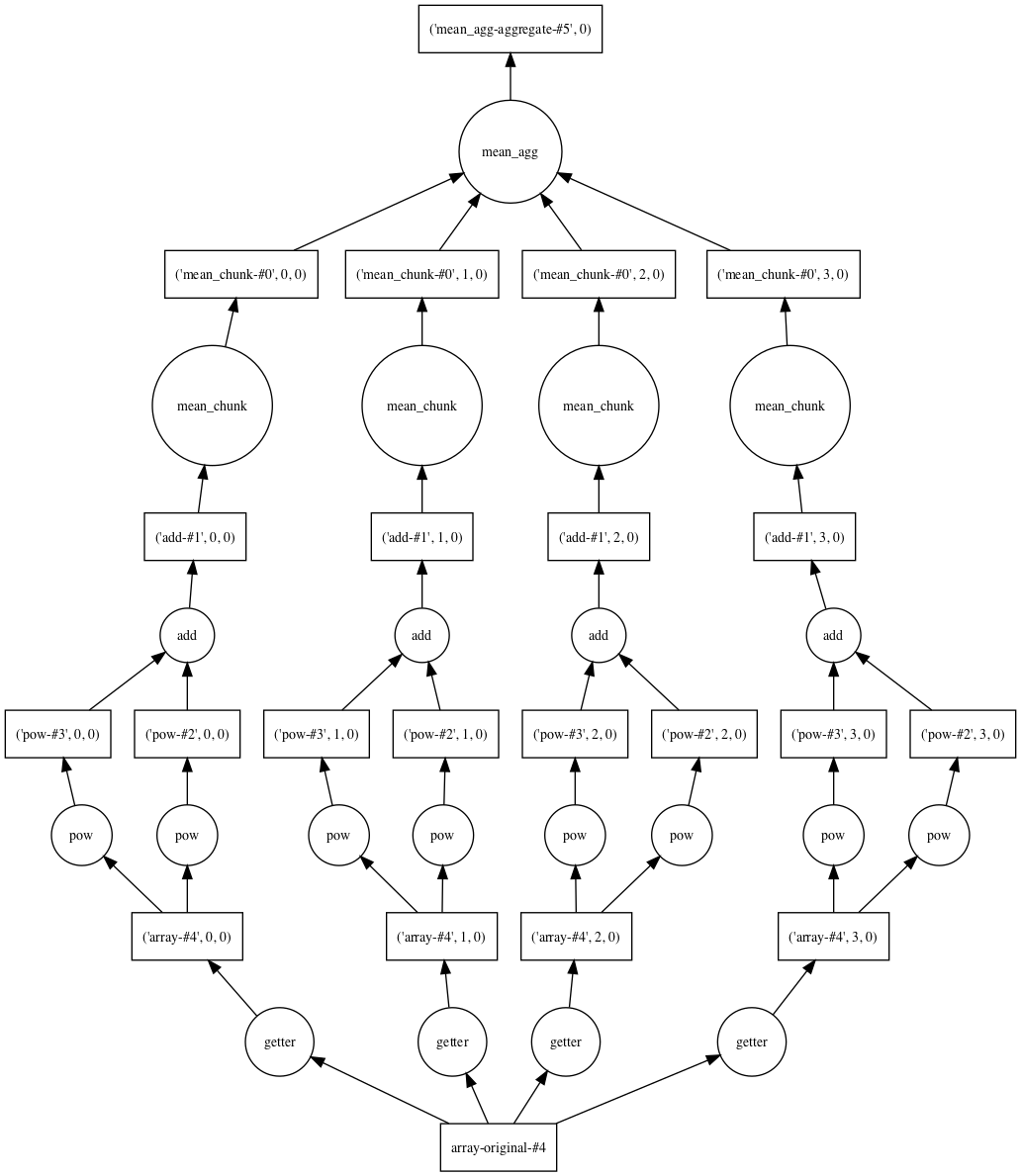
Now do the calculation¶
[10]:
with Profiler() as prof, ResourceProfiler(dt=0.1) as rprof,\
CacheProfiler() as cprof:
answer = result.compute()
Visualize the cpu, memory and cache for the 4 threads
[11]:
visualize([prof, rprof,cprof], min_border_top=15, min_border_bottom=15)
[11]:
You can evaluate your own functions on dask arrays¶
If your functons release the GIL, you can get multithreaded computation using dask.delayed